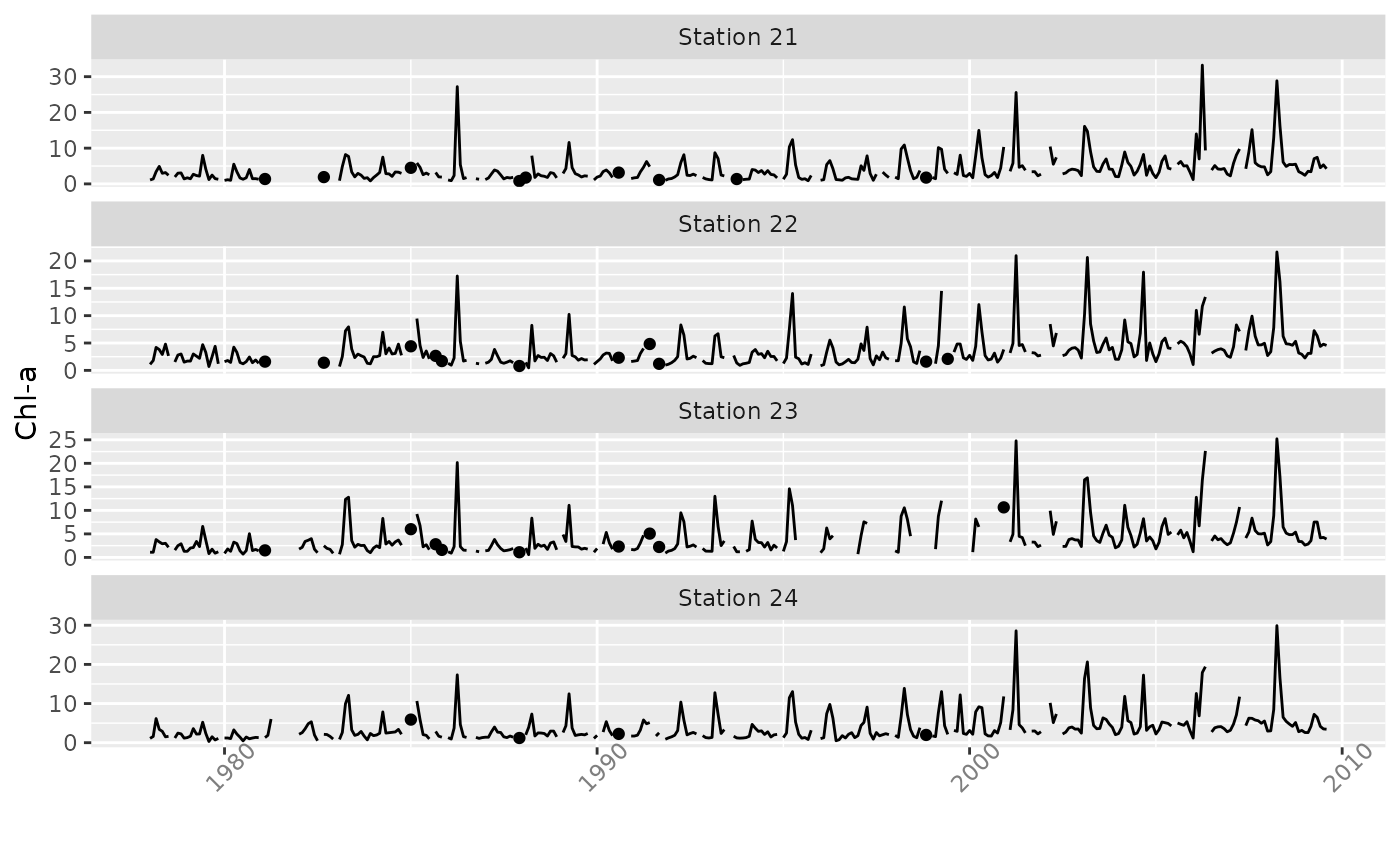
Creates line plot of vector or matrix time series, including any data surrounded by NAs as additional points.
plotTs(
x,
dot.size = 1,
xlab = NULL,
ylab = NULL,
strip.labels = colnames(x),
...
)Arguments
- x
matrix or vector time series
- dot.size
size of dots representing isolated data points
- xlab
optional x-axis label
- ylab
optional y-axis label
- strip.labels
labels for individual time series plots
- ...
additional options
Value
A plot or plots and corresponding object of class “ggplot”.
Details
The basic time series line plot ignores data points that are adjacent to
missing data, i.e., not directly connected to other observations. This can
lead to an uninformative plot when there are many missing data. If one
includes both a point and line plot, the resulting graph can be cluttered
and difficult to decipher. plotTs plots only isolated points as well
as lines joining adjacent observations.
Options are passed to the underlying facet_wrap function in
ggplot2. The main ones of interest are ncol for setting the
number of plotting columns and scales = "free_y" for allowing the y
scales of the different plots to be independent.