Generating Hypsography
Jemma Stachelek
2022-06-22
hypsography.RmdIntroduction
The General Lake Model (GLM) requires information on the depth-area relationship for a given lake. In GLM this is referred to as hypsography (rather than bathymetry) and has the following description in the glmtools documentation for get_hypsography:
Example
Let’s start by investigating (reverse-engineering) the hypsography provided for Sparkling Lake (Wisconsin) in the run_example_sim function.
sim_folder <- run_example_sim("inst/extdata", verbose = FALSE)
nml_file <- file.path("inst/extdata", "glm2.nml")
nml <- read_nml(nml_file)
hp <- get_hypsography(file = nml_file)[,2:1]Calculate volume from hypsographic profile
hp_diff <- cbind(c(diff(hp$depths), 0), hp$areas)
calc_layer_vol <- function(hp_diff, i){
top_area <- hp_diff[i, 1]
bot_area <- hp_diff[i + 1, 2]
# volume of a layer approximated as a truncated cone
(hp_diff[i, 1] / 3) * (top_area + bot_area + sqrt(top_area * bot_area))
}
vol_layers <- sapply(seq_len(nrow(hp_diff) - 1),
function(i) calc_layer_vol(hp_diff, i))
(vol_estimated <- sum(vol_layers))## [1] 153146165Approximate hypsographic profile from max depth and surface area
(nlayers <- nrow(hp))## [1] 15
(max_depth <- tail(hp$depths, 1))## [1] 24.9936
(surface_area <- head(hp$areas, 1))## [1] 39581170
res <- suppressWarnings(
glmutils::generate_hypsography(nlayers, max_depth, surface_area))
plot(res[,c("area", "depth_linear")],
ylim = c(max(res$depth_linear), min(res$depth_linear)),
ylab = "depth")
points(res[,c("area", "depth_ellipsoid")], col = "red", pch = 19)
legend("topleft", legend = c("linear", "ellipsoid"), col = c("black", "red"),
pch = c(21, 19))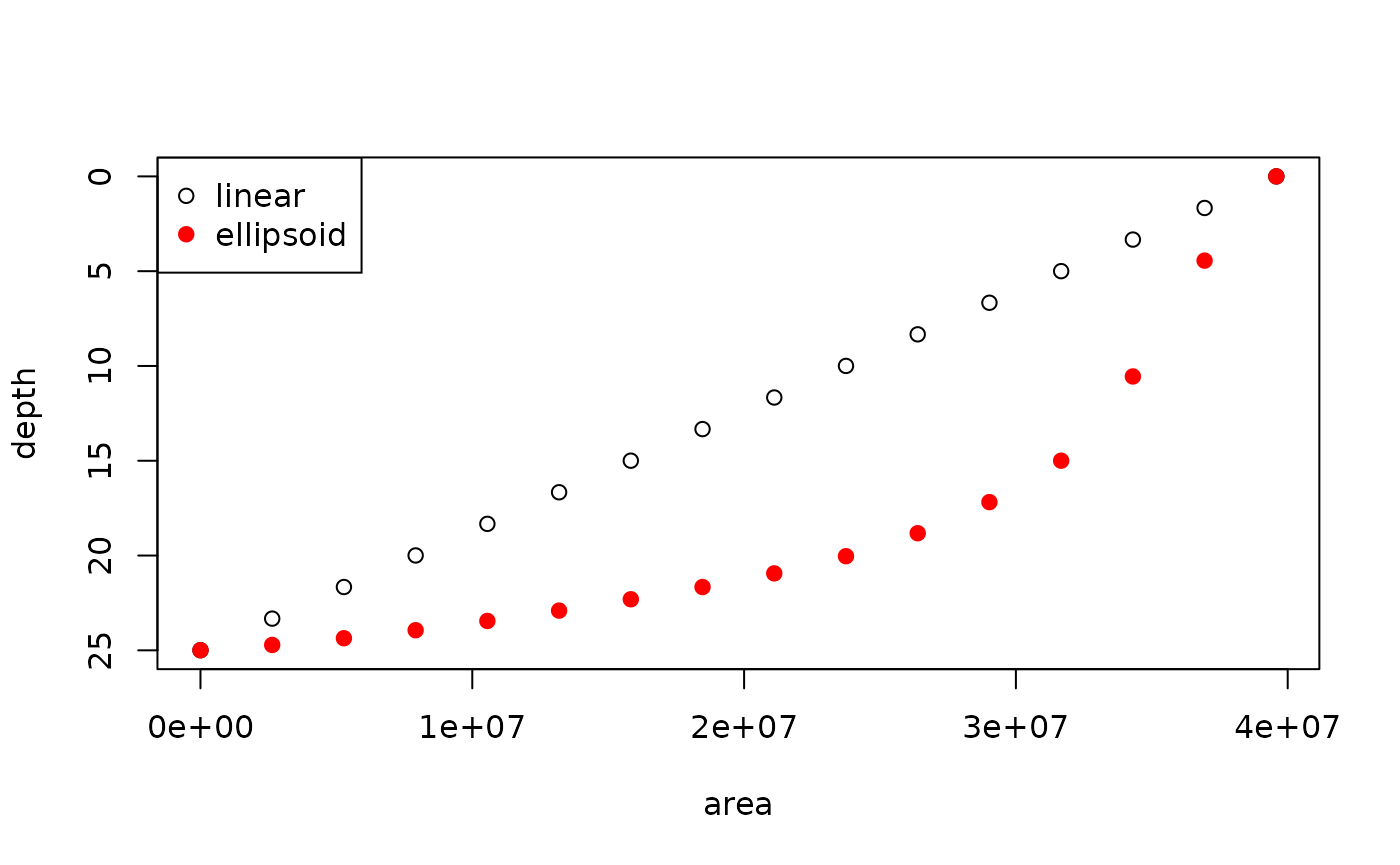
Verify max depth
# Get max depth
tail(hp, 1)| areas | depths | |
|---|---|---|
| 15 | 0 | 24.9936 |
This value matches the max depth reported by the Wisconsin DNR.
Verify dimensions
get_nml_value(nml, "bsn_len")## [1] 9500
get_nml_value(nml, "bsn_wid")## [1] 7400
library(nhdR)
# Get NHD Waterbody Polygons
# nhd_plus_get(vpu = 7, "NHDSnapshot")
dt <- nhd_plus_load(vpu = 7, "NHDSnapshot", "NHDWaterbody")
spark_outline <- dt[grep("Sparkling", dt$GNIS_NAME),]
plot(spark_outline$geometry, main = "Sparkling Lake")
suppressPackageStartupMessages(library(lakemorpho))
library(raster)
# Get length and width
spark_outline_sp <- as(spark_outline, "Spatial")
r <- rasterize(spark_outline_sp, raster(spark_outline_sp))
spark_outline_sp <- spTransform(spark_outline_sp, CRS("+proj=utm +zone=10 +datum=WGS84"))
r <- projectRaster(r, crs = "+proj=utm +zone=10 +datum=WGS84")
spark_lm <- lakeSurroundTopo(spark_outline_sp, r)
calcLakeMetrics(spark_lm, bearing = 45, pointDens = 250)