Check for Braiding in a River Network
Detects braiding (multiple flow channels between two locations) within a river network object. Braiding can either be checked for in the route between two segments, or in the river network as a whole.
checkbraided(rivers, startseg = NULL, endseg = NULL, progress = TRUE)
Arguments
| rivers | The river network object to check. |
|---|---|
| startseg | Starting segment of a route to investigate. If this and
|
| endseg | Starting segment of a route to investigate. If this and
|
| progress | Whether to show the progress bar. Defaults to |
Note
This function is called within cleanup, which is recommended in most cases.
Examples
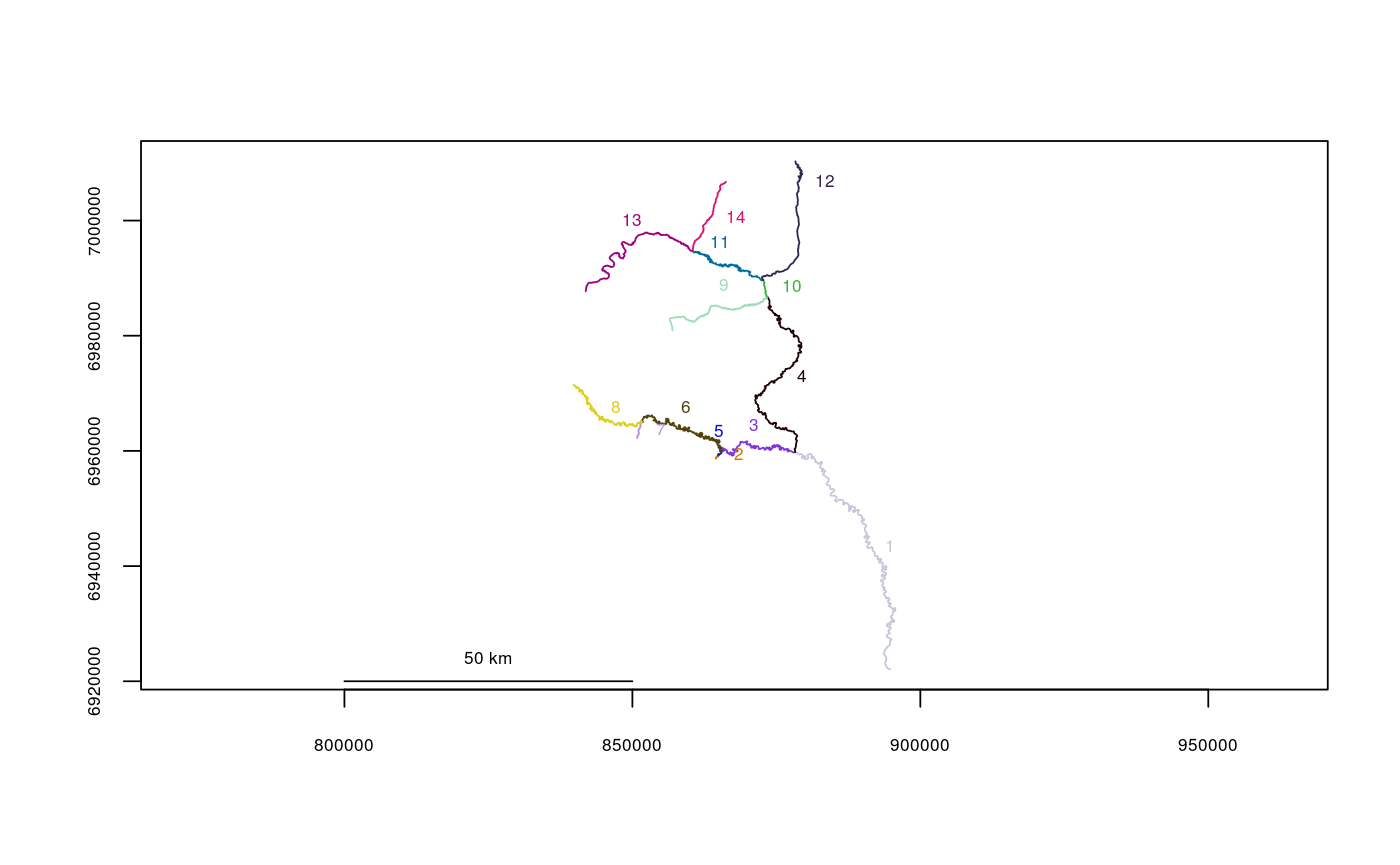
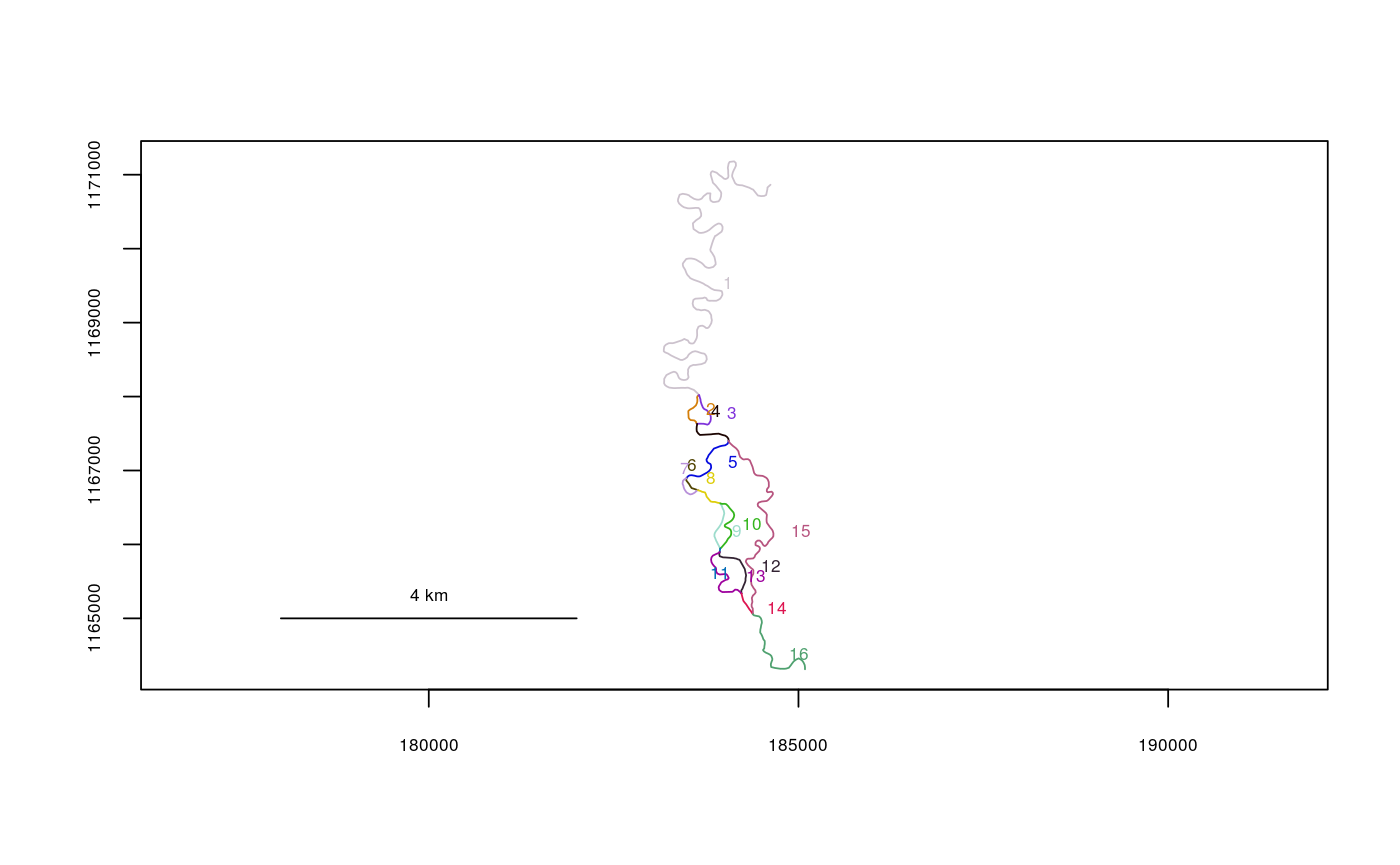
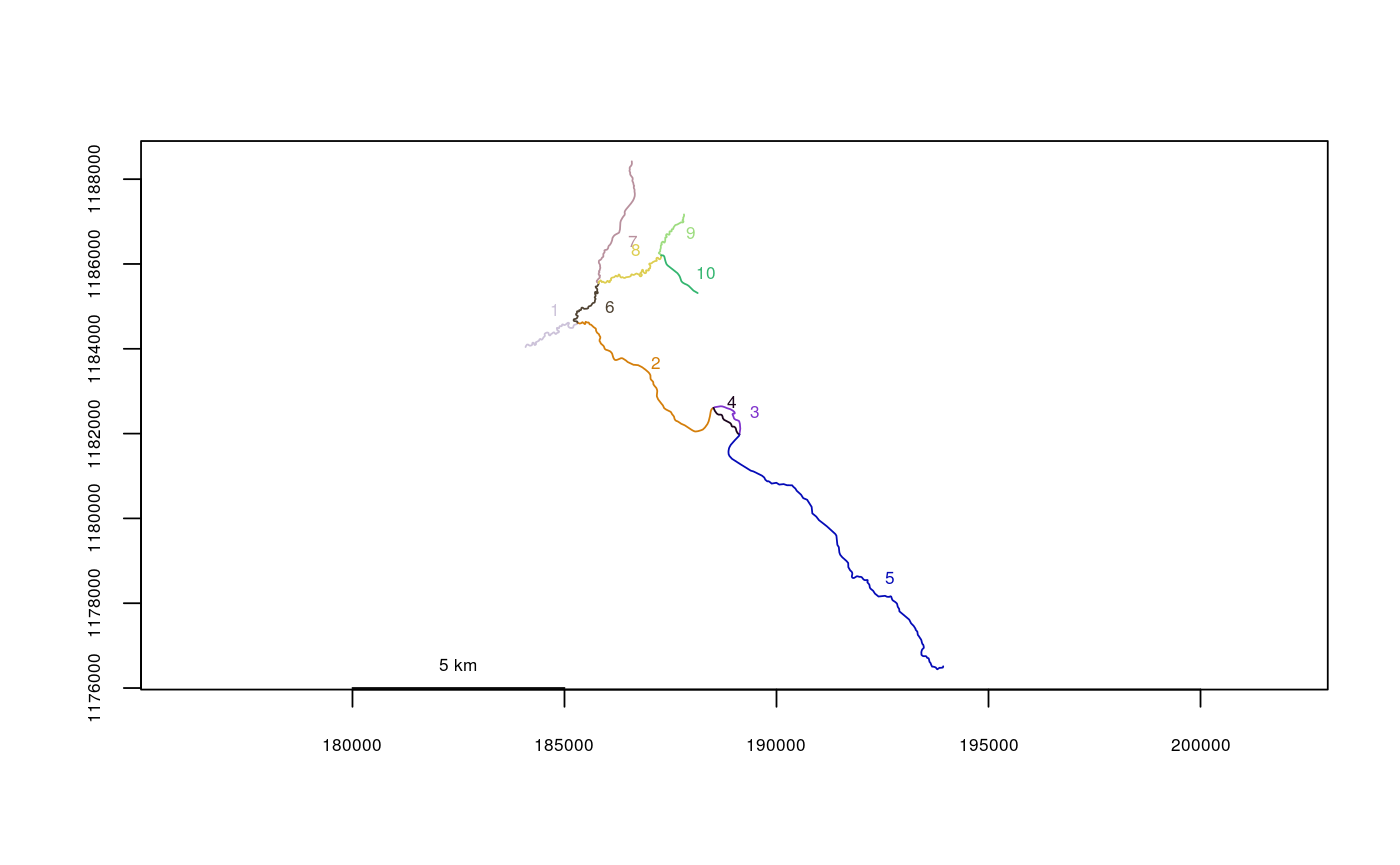
data(Gulk) plot(x=Gulk)checkbraided(rivers=Gulk)#> | | | 0% | |================== | 25% | |========================== | 38% | |=================================== | 50% | |============================================ | 62% | |==================================================== | 75% | |============================================================= | 88% | |======================================================================| 100% #> No braiding detected in river network.data(KilleyW) plot(x=KilleyW)checkbraided(rivers=KilleyW)#> | | | 0% | |======================================================================| 100% #> Braiding detected in river network. Distance measurements may be inaccurate.#>plot(x=Kenai3.subset)checkbraided(startseg=1, endseg=7, rivers=Kenai3.subset)#> No braiding detected between segments.checkbraided(startseg=1, endseg=5, rivers=Kenai3.subset)#> Braiding detected between segments. Distance measurements may be inaccurate.